CRASH COURSE ON NANOPORE SEQUENCING FOR MICROBIAL ECOLOGY– Rome edition
11- 14 March 2024, CNR-IRSA, Roma, Italy

The 13th edition of the International Course in Microbial Ecology -ICME13 focuses on the Nanopore sequencing techniques and data analysis for microbial ecologists. It is a practical course addressed to maximum 25 PhD students and early career researchers. The course includes the following activities:
- Practical experience in sequencing using Nanopore technology (MinIon)
- Bioinformatic analysis of Nanopore sequencing data
During the course, students will generate and analyze metagenomic data from DNA samples previously extracted and tested for the training activities. The course is hand-on training with a clear practical setting, preceded by theoretical activities. Online bioinformatic and ad hoc customized pipelines will be presented. Participants will have to introduce themselves with a 3 minutes presentation, including their main research topic.
Course structure:
- Day 1, Monday 11 March 2024: 14:00- 19:00. Opening, students presentations at the CNR-Headquarter. Social dinner
- Day 2, Tuesday 12 March 2024: 8:00- 18:00. Transfer to CNR-IRSA, Montelibretti. Practical laboratory activies, theoretical lessons
- Day 3, Wednesday 13 March 2024: 8:00- 18:00. Transfer to CNR-IRSA, Montelibretti. Practical laboratory activies, theoretical lessons
- Day 4, Thursday 14 March 2024: 9:00- 17:00. Transfer to CNR-IRSA, Montelibretti. Plenary lecture, presentation of the results by participants, closing ceremony
Application and registration fee
Participants are selected based on their CV and motivation letter. No prior knowledge with bioinformatics is needed but familiarity with terminology and concepts is preferred. Application can be done by filling up the ICME13 -registration form (Application on line botton). More information can be requested by email at e-mail (icme13_roma@microbeco.org).
The registration fee is 350 euro/person and it includes the full participation to all the activities, and lunches, coffee breaks and the social evening and public transports ticket to reach the CNR-IRSA Montelibretti. Accommodation and travel costs are excluded. At the moment no travel grants are available. N.B: The use of a personal computer with adapter socket and, possibly, extension cable is required. Selected participants need to prepare 2-3 minute presentation about his/her research.
Important dates
5 February – Application deadline
By this date, applicants should send their CV Applications should send max 1 page CV and 1/2 page motivation letter or apply directly using the In the motivation letter the candidate should explain: a) the reason to attend the crash course in Nanopore sequencing for microbial ecology; b) any experience in bioinformatics; CV and motivation letter are the basis for selection. You should convince the committee you need the course and you can contribute to its successful realization.
12 February – Notification of acceptance
Candidates will be notified of being selected and will have to accept and confirm their participation. The waiting list scrolls down until 19 February.
4 March – Deadline payment for the registration fees
The 350 euro registration fee includes all didactical materials, with access to a shared folder, lunches, coffee breaks and the social dinner on Monday. Regional train tickets to and back from the CNR-IRSA at Montelibretti are included too. Accommodation and travel expenses are excluded. A list of suggested accommodation is provided to help for a better arrangement.
11 March – 14:00 – Beginning of the course at the CNR-headquarter; Piazzale Aldo Moro, 7 – Roma
The course will start on Monday 11/03 at 14.00. Participants should prepare 2-3 minute presentation about his/her research activities.
Invited speakers and instructors

Martina Cappelletti is Associate Professor at the University of Bologna (Department of Pharmacy and Biotechnology) teaching courses on Microbial Genomics and Molecular Microbiology. She leads the Molecular Environmental Microbiology group (MEMlab) focused on the study of genomic, molecular, and biotechnological aspects of microbial interactions with organic and inorganic substrates/compounds and bacterial response to environmental stresses.

Ester Eckert is a researcher at the CNR-Institute of Water research in Verbania. Her scientific activity comprises a broad range of topics from animal microbiomes, over antibiotic resistance genes to meiofauna in mainly aquatic environments. She uses a multidisciplinary approach for investigating how species interactions on multiple trophic levels influences the persistence of antibiotic resistance genes and bacteria in the environment.

Grazia Marina Quero is a researcher at the CNR Institute of Marine Biological Resources and Biotechnology. She works on the ecological role of prokaryotes in coastal and deep marine ecosystems, the anthropogenic impact on biodiversity and the functioning of aquatic microbial communities, and the microbiome associated with different marine organisms, including invertebrates and fish.

Andrea Firrincieli received his PhD in Science, Technology, and Biotechnology at the University of Viterbo in 2017. Since then, Andrea has worked in the microbial genomic and microbial ecology fields focusing on the study of microbes and microbial communities inhabiting natural and artificial environments. His area of expertise falls in the next- and third-generation sequencing data analysis, comparative genome analyses, and phylogenomics. His current work at the University of Viterbo focuses on the study of fungi-bacteria and plant-microbes interactions in soil bioremediation and phytoremediation.

Tomasa Sbaffi is a post-doc at the Microbial Ecology Group in Verbania. Her research activities regards microbial ecology/evolution of prokaryotes and microbial eukaryotes in natural/controlled environment, statistical methods to analyse community structure, synthetic communities, ecological functions, public good dynamics, and bioinformatics with a focus on environmental antimicrobial resistance.

Davide Corso is a bioinformatician PostDoc in the laboratory of Donato Giovannelli and Angela Cordone at the University of Naples Federico II. His interests in bioinformatics have expanded over time and he continues studying and increasing his knowledge of various biological applications. His current topics regard microbial ecology, metagenomics, earth system science, and astrobiology. Hs is also involved in projects related to developing new bioinformatics tools or data analysis for example, in the identification of antibiotic-resistance genes.

Marco Basili is a PhD student of the University of Bologna in collaboration with the IRBIM-CNR (Ancona).
In his PhD project he is performing bioinformatics analysis on microbial communities from several types of fish and environmental samples; For these analyzes he is currently using 16S rRNA Gene Metabarcoding and Metagenomics with focus on Antimicrobial resistance genes.
Organizing Committee
Several researchers and communicators contributed to the organization of the course: Bruna Matturro ,Stefano Amalfitano , Bruno Benedetti, Valerio Bocci, Alessio Massimi – CNR-IRSA Roma; Gian Marco Luna, Grazia Quero, Manuela Coci – CNR-IRBIM- Ancona | Elena Panariello – MicrobEco | Luigi Gallucci – Max Planck Institute for Marine Microbiology & MicrobEco | Ester Eckert – CNR-IRSA-Verbania .
Location and practical information
The venue
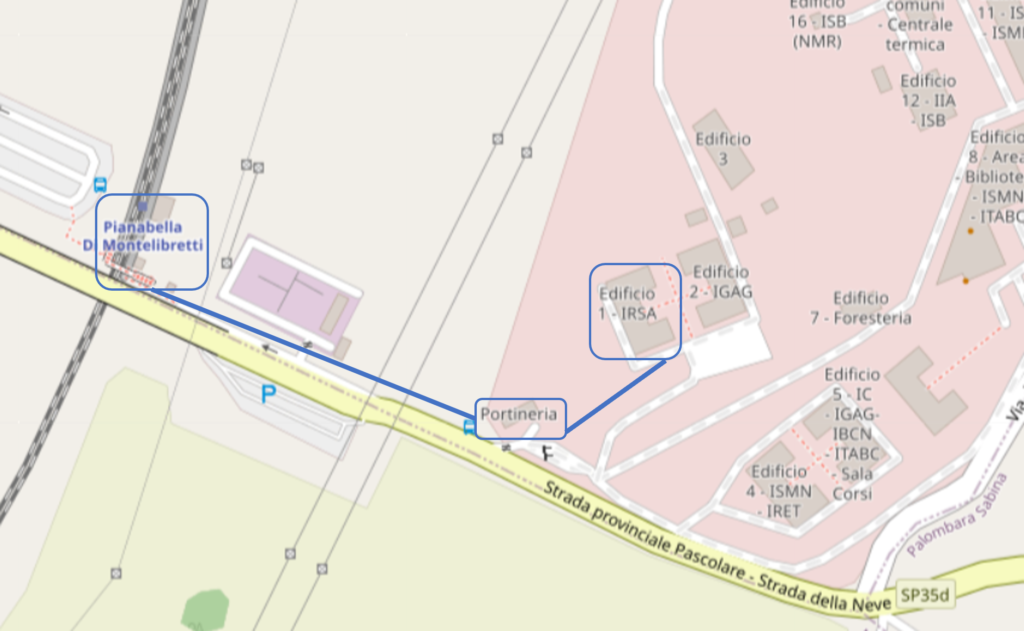
The course will take place in two different locations. The opening on Monday 11 March 2024 will take place at the CNR-headquarter. From Tuesday to Thursday the course will be held at the CNR–IRSA inside the RM1- Montelibretti Research Area (AdR RM1), located at approximately 30 km from Rome, in the North-Est region. Participants will receive a daily train ticket to go and come from the CNR-IRSA. The nearest train stop is “PIANABELLA DI MONTELIBRETTI”, a few steps from the main entrance of the Area. Starting train stations in Rome are: Roma Nomentana and Roma Tiburtina. Regional trains from Roma Tiburtina or Nomentana to Piana Bella di Montelibretti last 30 minutes and go approximately every 15 minutes. More detailed information will be provided at the course. Once at the entrance of the research area, it is necessary that visitors present themselves at the reception, showing their identity card. They will receive a special badge and a form to be filled out by the local host of the CNR. The CNR-IRSA building is just a 5-minute walk from the reception as indicated in the map.


Roma by plane
Fiumicino – “Leonardo da Vinci” airport of Rome is the biggest airport in Italy. It has several international low-cost and full-services airlines. It is located about 25 km southwest of the city of Rome. The Leonardo Express offers direct connection to the Termini train station for 11 euro. Tickets can be bought at the airport through ticket machine, from the ticket/info stands or on line through the Trenitalia Website (using “Fiumicino Aeroporto” and “Roma Termini”).
Rome-Ciampino Airport is a very small airport about 13 km distant from Rome. It serves various low-cost airlines, including Ryanair and Wizzair. Private buses offer continuous service to and from Termini and Tiburtina stations
Roma by train
Rome is well connected to the major Italian cities by both regional and high-speed trains. Roma Termini is the main station of the capital, providing regional, national and international connections. Direct services are provided by Trenitalia, through Frecciarossa and Frecciargento, and by ITALO, thanks to their new high-speed trains, allowing people to travel in the city in a fast, easy and comfortable way.
Tips for accommodation in Rome
Roma offers hundreds of accommodations for all types of budget, but is also very big and chaotic. We therefore suggest finding accommodation at a walking distance from the station Roma Tiburtina. A convenient point of reference is Piazza Bologna in the Nomentano district. In Piazza Bologna, there is the metro stop of the B1 line (Blu line), a road junction of great importance. Line 90 connects it to Termini Station, as well as from Piazza Venezia or Via Nazionale (with bus 60). Below a map of the Metro in Rome linked to the wepage. Do no hesitate to ask suggestions by writing to icme13_rome@microbeco.org